Biomolecular design
DrugCLIP: Contrastive Protein-Molecule Representation Learning for Virtual Screening
Advisor: Prof. Wei-Ying Ma, Prof. YanYan Lan, Institute for AI Industry Research, Tsinghua University
Worked with Bowen Gao, Bo Qiang, Haichuan Tan, Yinjun Jia, Minsi Ren
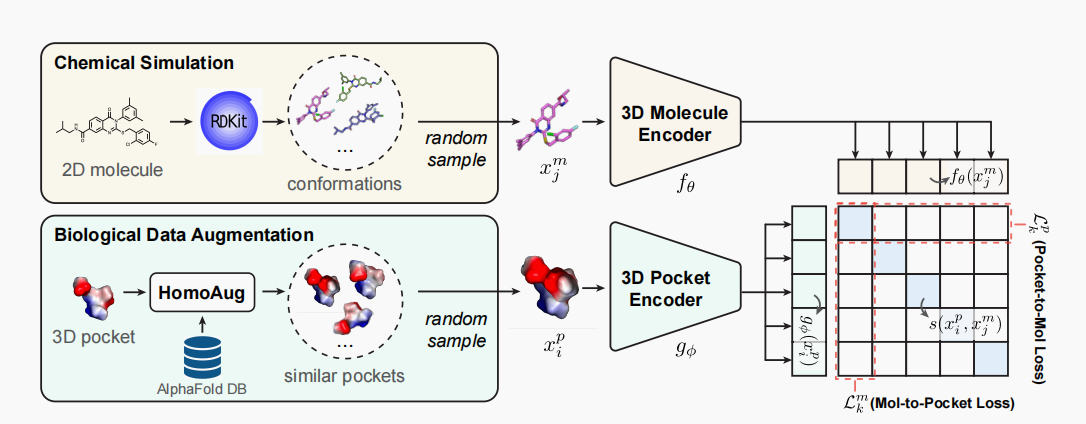
DrugCLIP is a cutting-edge approach to virtual screening. It reformulates virtual screening as a dense retrieval task and employs contrastive learning to align representations of binding protein pockets and molecules from a large quantity of pairwise data without explicit binding-affinity scores.[Read more]
Omics Data
Capricorn: a multi-view diffusion model for Hi-C contact matrix resolution enhancement
Advisor: Prof. William Stafford Noble and Prof. Sheng Wang, School of Computer Science & Engineering, University of Washington
Worked with Tangqi Fang, Yifeng Liu, Addie Woicik
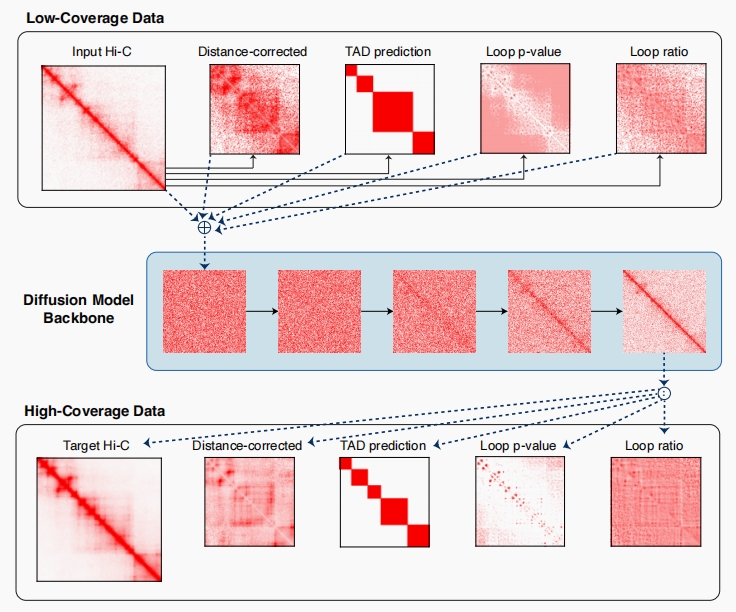
A tool for Hi-C resolution enhancement that incorporates chromatin features (like loops and TADs) as additional views of the input Hi-C contact matrix and leverages a diffusion probability model backbone to generate a high-resolution matrix. Capricorn is a powerful Hi-C resolution enhancement method that enables scientists to find chromatin features that cannot be identified in the low-coverage contact matrix. [Read more]
A tool for the analysis of time series Hi-C
Advisor: Prof. William Stafford Noble and Prof. Sheng Wang, School of Computer Science & Engineering, University of Washington
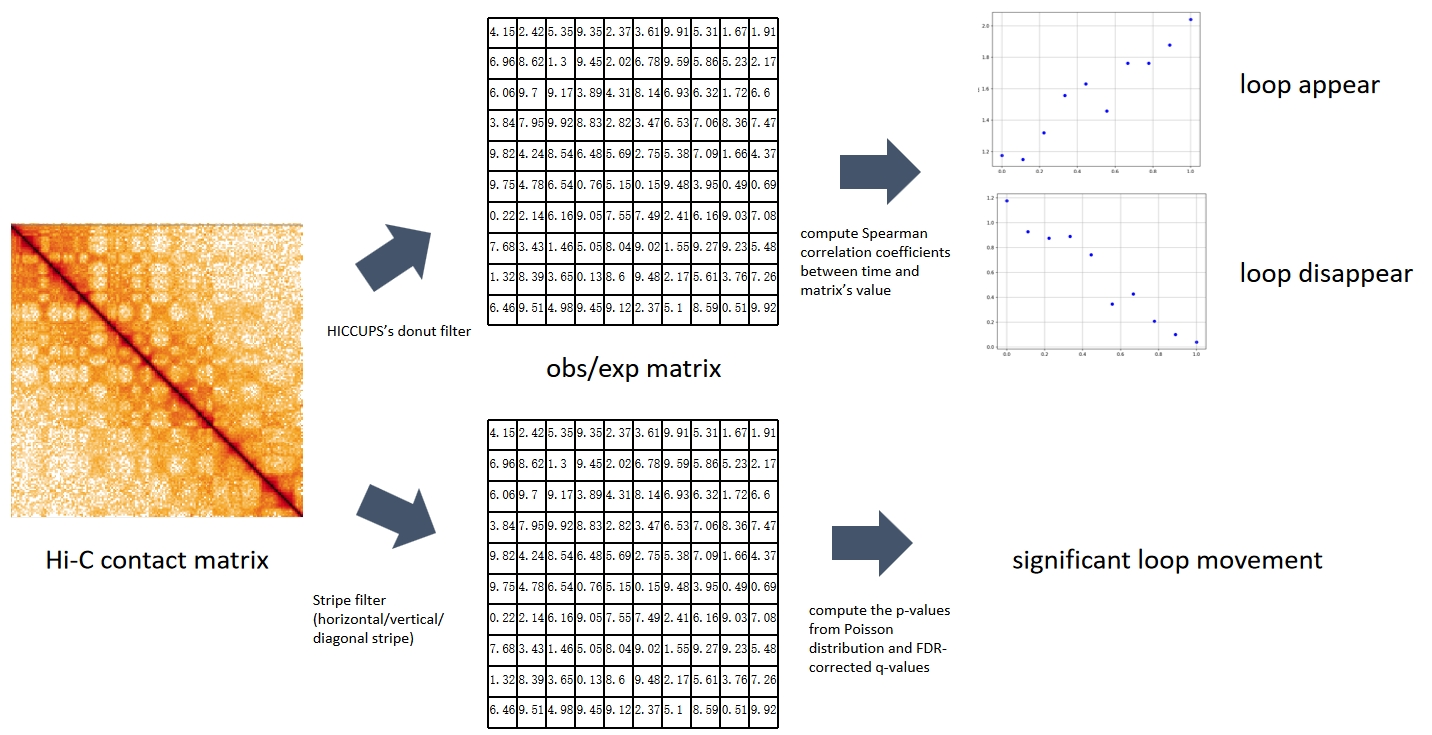
Changes in spatial genome architecture over time necessitate the joint temporal analysis of Hi-C. We developed a tool for Hi-C time series to study how chromatin structure changes over time in development, the cell cycle, and disease progression. We focus on loop dynamics and discover the appearance, disappearance, and movement patterns for the Hi-C contact map.[Read more]
Multiomics Integration Via Graph Learning
Advisor: Jianyang Zeng, Tsinghua University
Worked with Wenda Chu, Botian Wang, Sihang Zeng, Shiyu Zhao
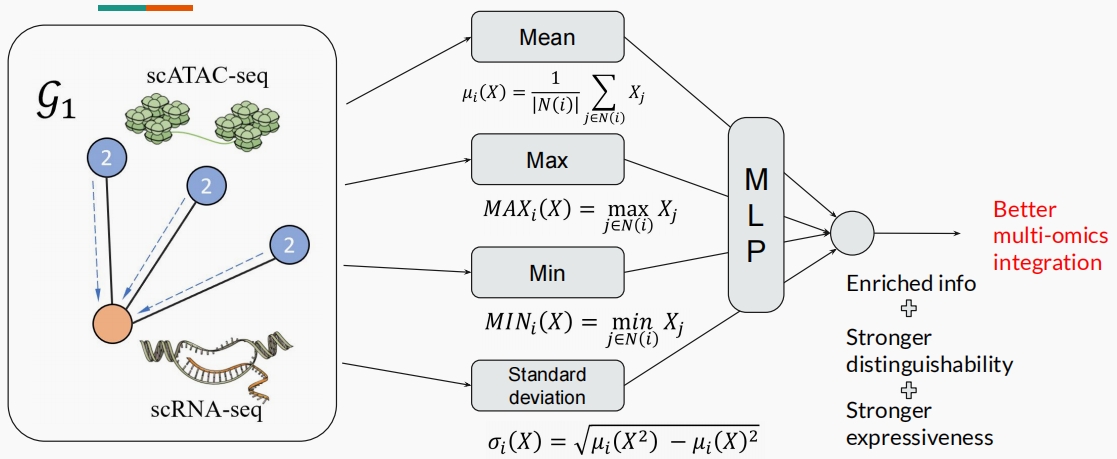
This project focuses on the integration of scRNA-seq and scATAC-seq data using a graph learning approach inspired by GLUE. The novelty of our method lies in decaying the weight of edges according to the distance of the ATAC peak from the TSS and using a set of independent aggregators to combine the messages in each GCN layer. Our method achieved comparative performance with GLUE on metrics and had the ability to identify more genes with regulatory relationships than GLUE. [Read more]
PPI Network Guided Driver Target Discovery
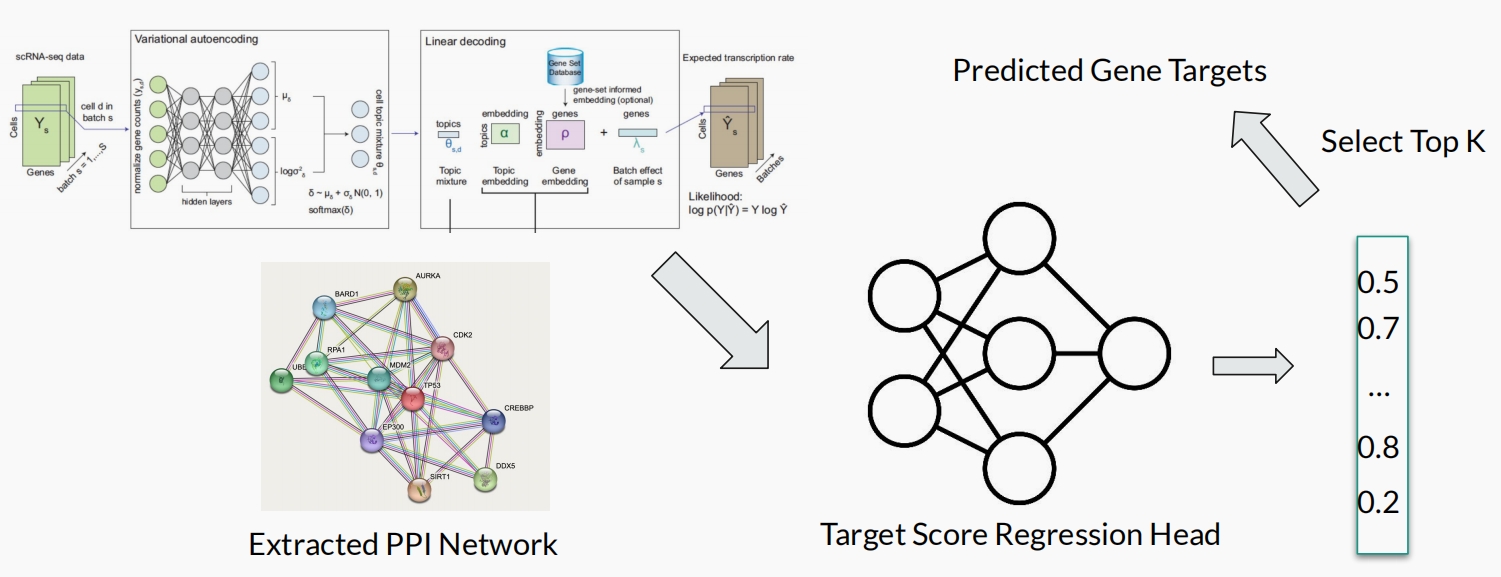
Advisor: Jianyang Zeng, Tsinghua University
Worked with Wenda Chu, Botian Wang, Sihang Zeng, Shiyu Zhao
We design a new method for driver target discovery using scRNA-seq data by introducing a protein-protein interaction (PPI) network to re-calulate regularization loss in the sc-ETM model and design a 2-layer MLP for target prediction. We achieved better cell clustering performance than sc-ETM and better driver target discovery performance. [Read more]
AI
Gomoku Genius: AI Search Strategies for Classic Board Game Mastery
Advisor: Prof. Mingsheng Long, Tsinghua University
This is a practical application of advanced AI search strategies in board games. We optimized Minimax for Tic-Tac-Toe. Applied Alpha-Beta Pruning to reduce search space. Implemented Truncated Search in Go-Moku, balancing time constraints with strategic depth using pattern evaluation. Conducted gameplay analysis comparing naive MCTS and Alpha-Beta. Enhanced MCTS with evaluation function, significantly improving board state assessments and decision-making.[Read more]
Text Sentimental Classification Based on the Twitter Dataset
Advisor: Prof. Mingsheng Long, Tsinghua University
In this project, we focus on comparing a variety of models using a Twitter sentiment analysis dataset. We utilized TF-IDF methods for feature extraction and implemented a range of machine learning models, including Decision Trees, Random Forests, Multilayer Perceptrons (MLPs), ResNet, and BERT, to conduct nuanced sentiment analysis.[Read more]